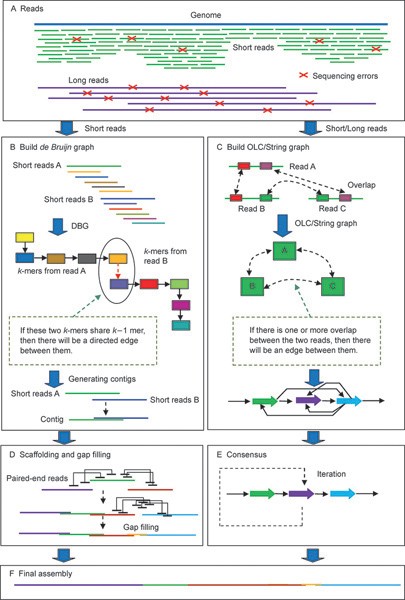
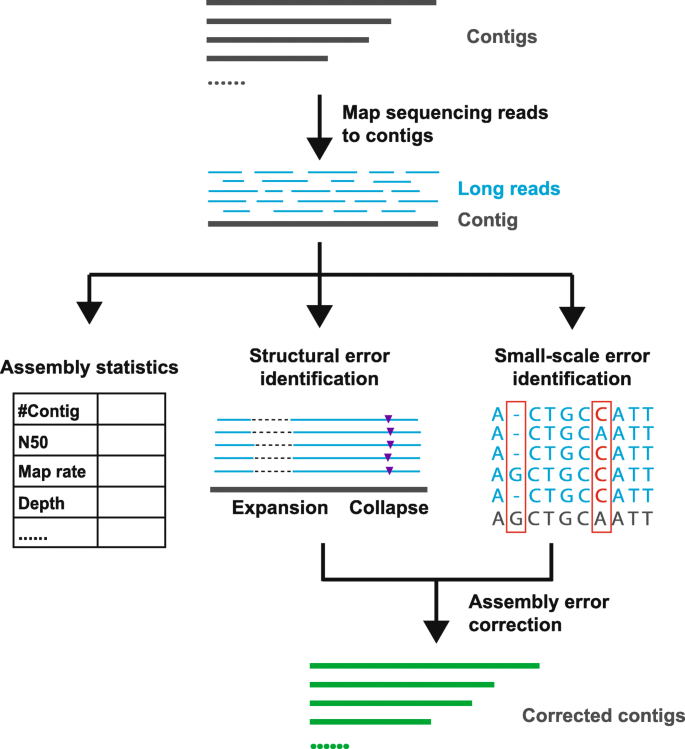
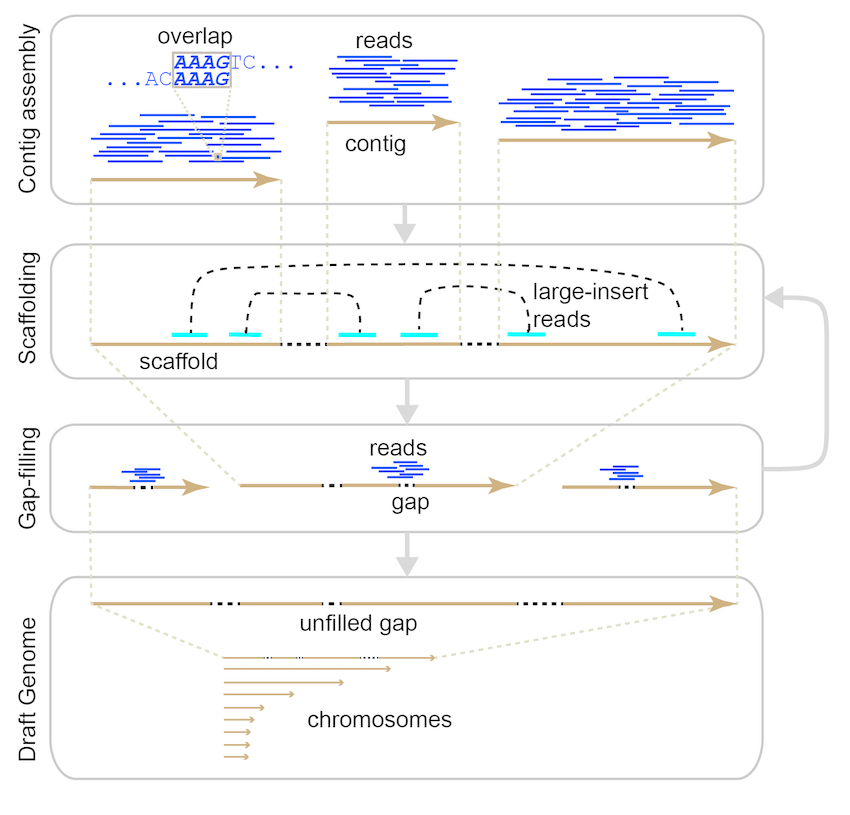
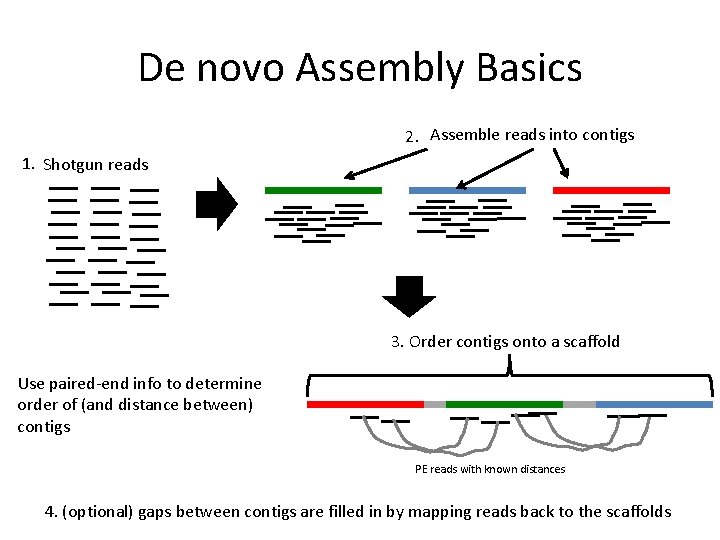
A) Scheme of the de novo based pipeline to identify possible spliced... | Download Scientific Diagram

Tools and Strategies for Long-Read Sequencing and De Novo Assembly of Plant Genomes: Trends in Plant Science
Selecting Superior De Novo Transcriptome Assemblies: Lessons Learned by Leveraging the Best Plant Genome
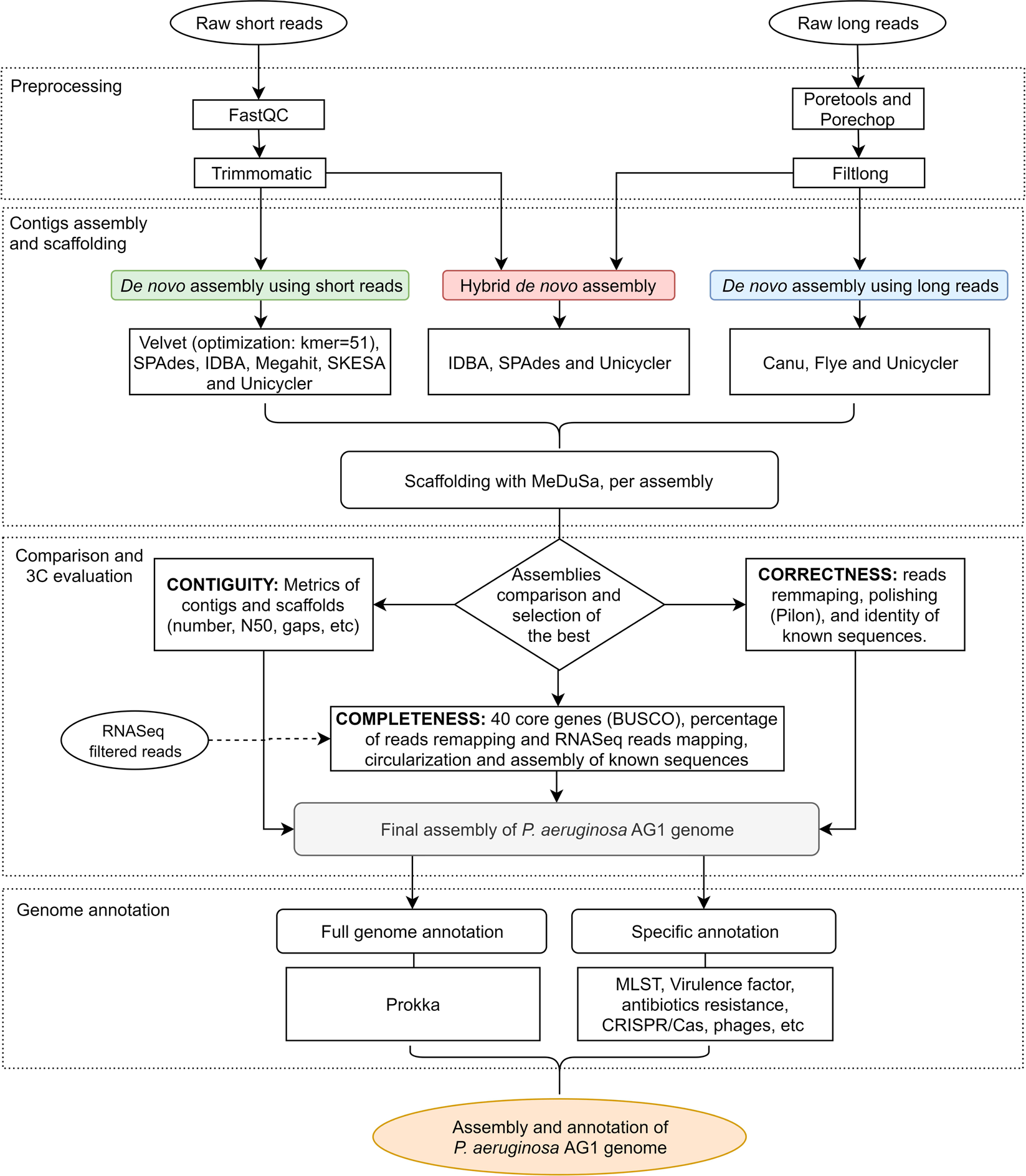
High quality 3C de novo assembly and annotation of a multidrug resistant ST-111 Pseudomonas aeruginosa genome: Benchmark of hybrid and non-hybrid assemblers | Scientific Reports

Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions | Nature Communications

Rapid hybrid de novo assembly of a microbial genome using only short reads: Corynebacterium pseudotuberculosis I19 as a case study - ScienceDirect

Gene Mapping | Genome Assembly and Annotation – 1010Genome | Quality NGS Bioinformatics Data Analysis Services

NT3-chitosan enables de novo regeneration and functional recovery in monkeys after spinal cord injury | PNAS
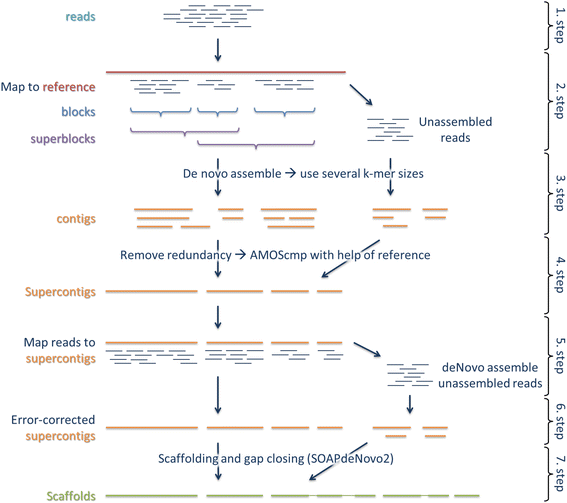
Reference-guided de novo assembly approach improves genome reconstruction for related species | BMC Bioinformatics | Full Text

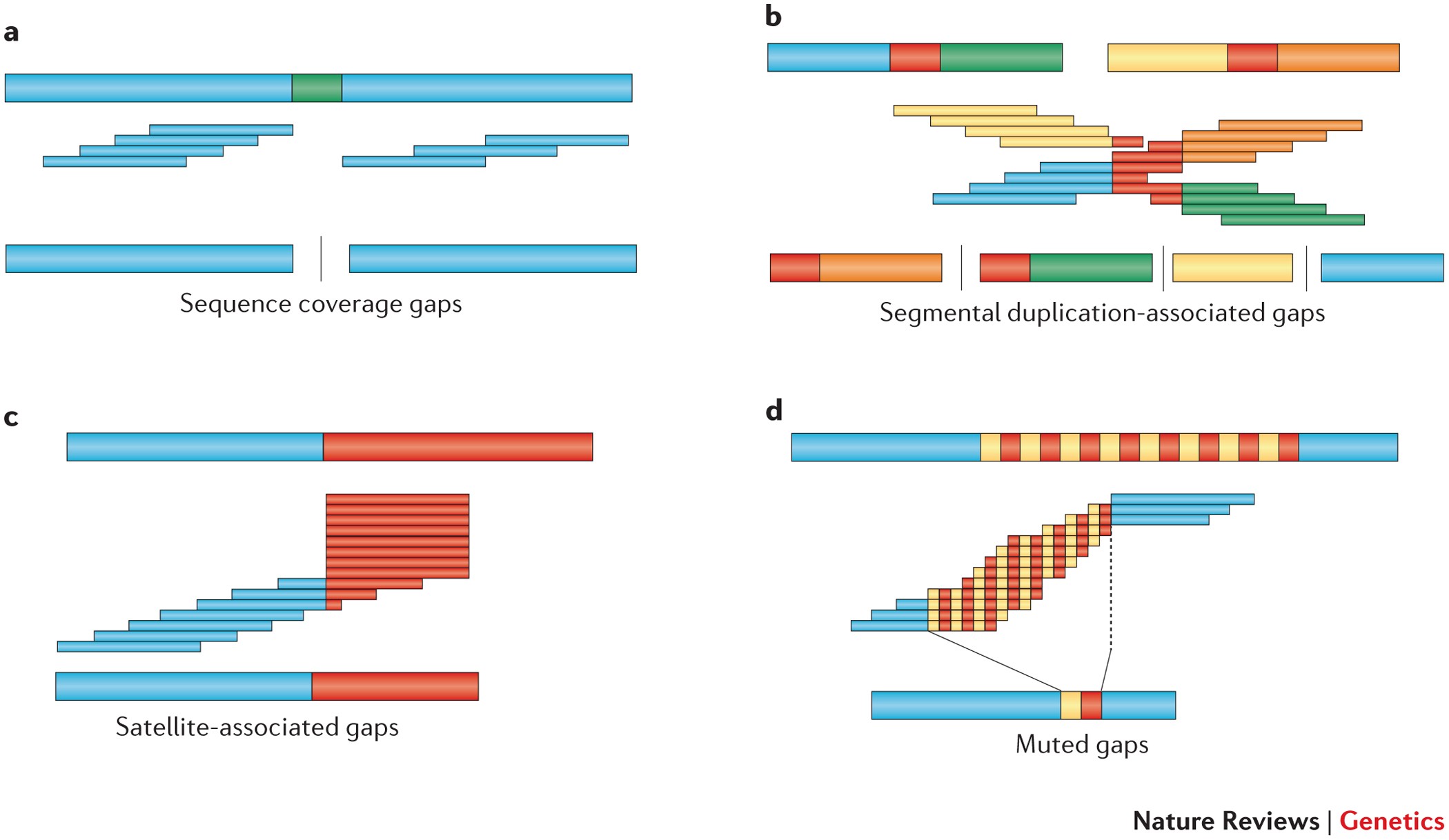
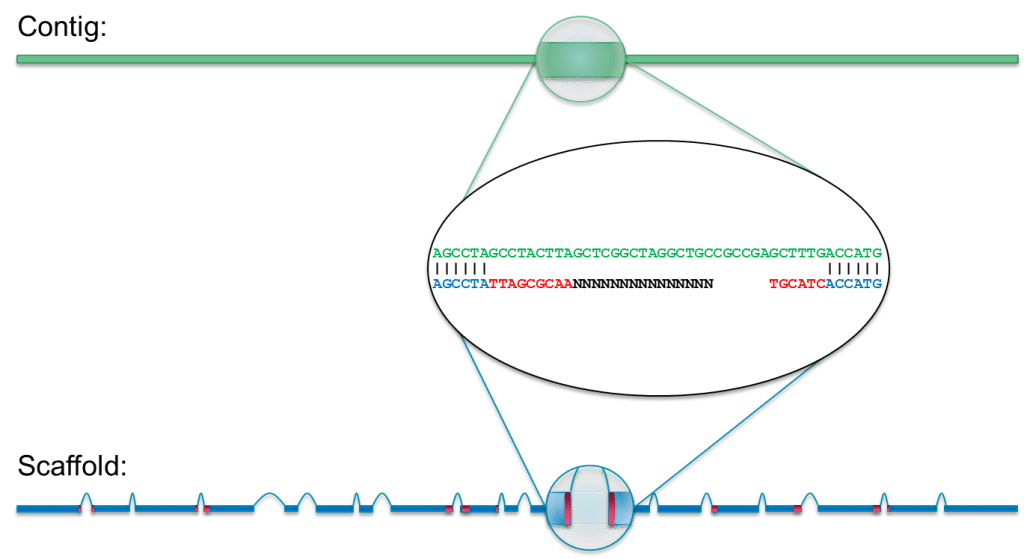
Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird‐of‐paradise - Peona - 2021 - Molecular Ecology Resources - Wiley Online Library

Protein de novo sequencing by top-down and middle-down MS/MS: Limitations imposed by mass measurement accuracy and gaps in sequence coverage - ScienceDirect
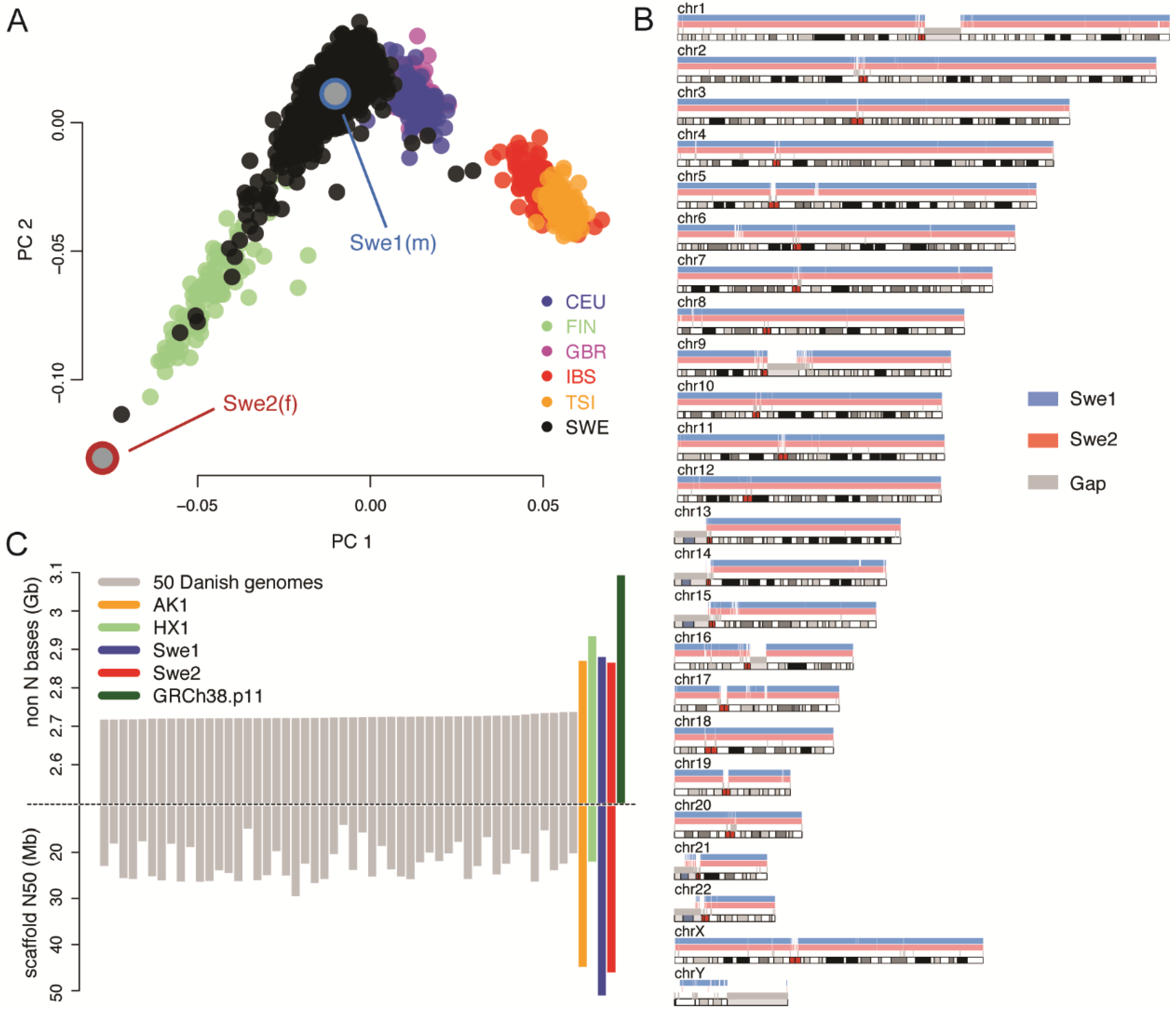
Genes | Free Full-Text | De Novo Assembly of Two Swedish Genomes Reveals Missing Segments from the Human GRCh38 Reference and Improves Variant Calling of Population-Scale Sequencing Data | HTML

The Kinetics of Deregulation of Expression by de Novo Methylation of the H19 Imprinting Control Region in Cancer Cells | Cancer Research
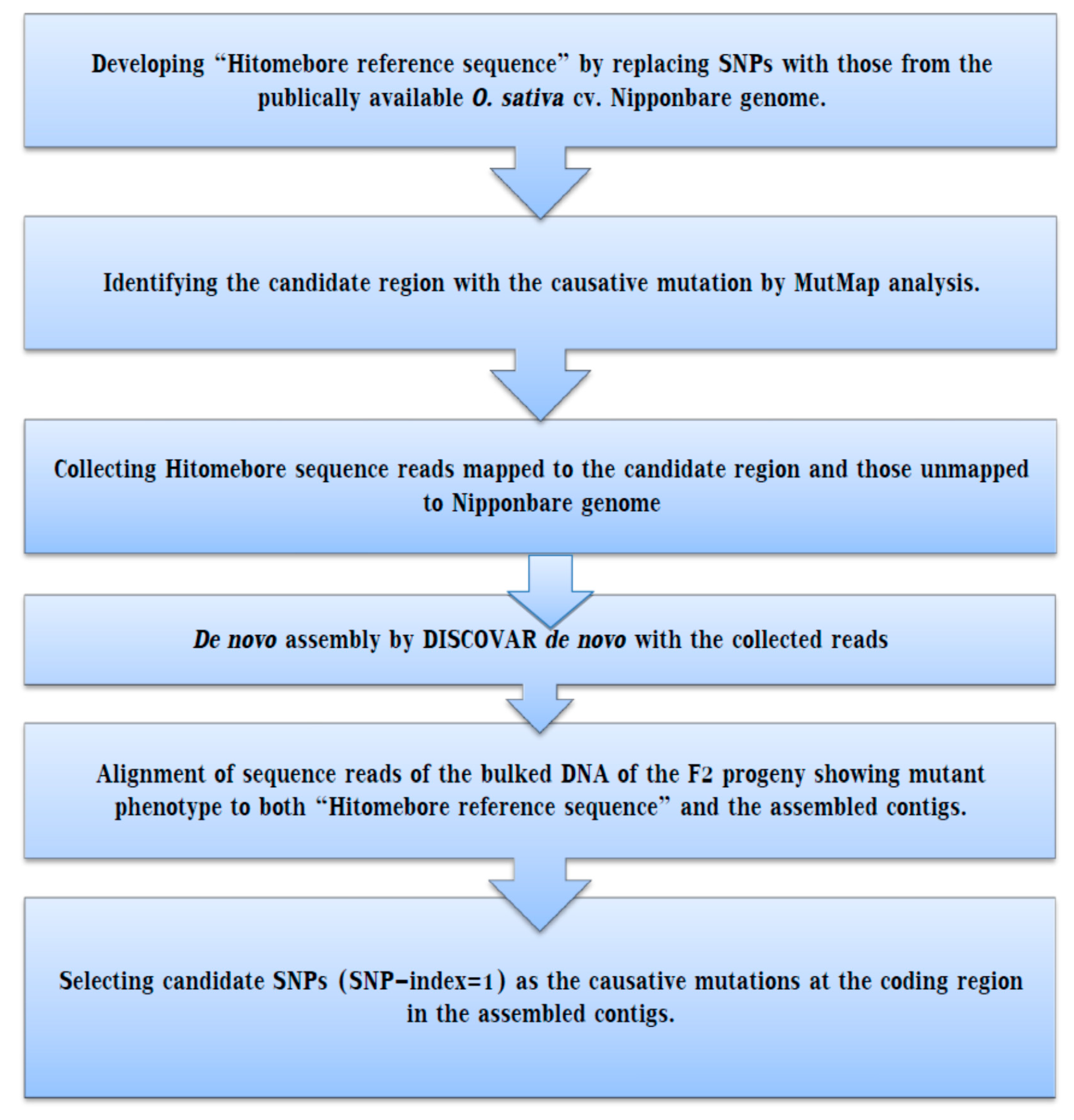
IJMS | Free Full-Text | Whole Genome Resequencing from Bulked Populations as a Rapid QTL and Gene Identification Method in Rice | HTML

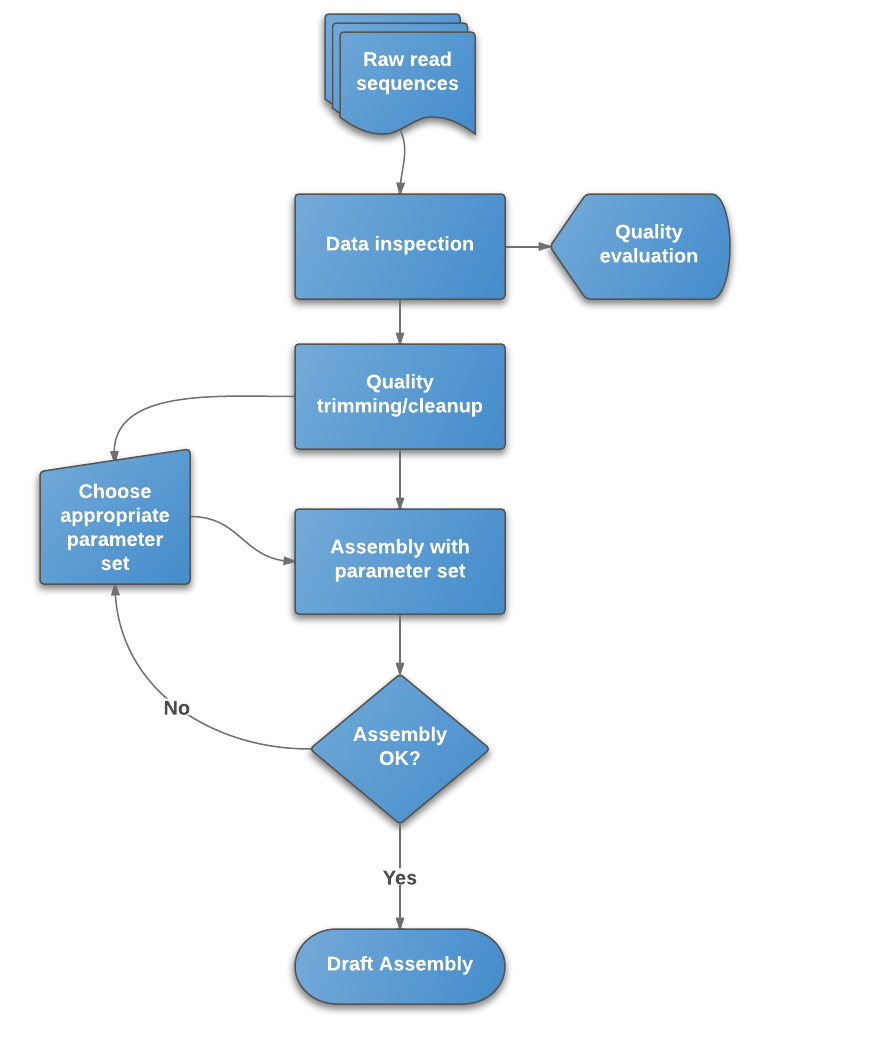

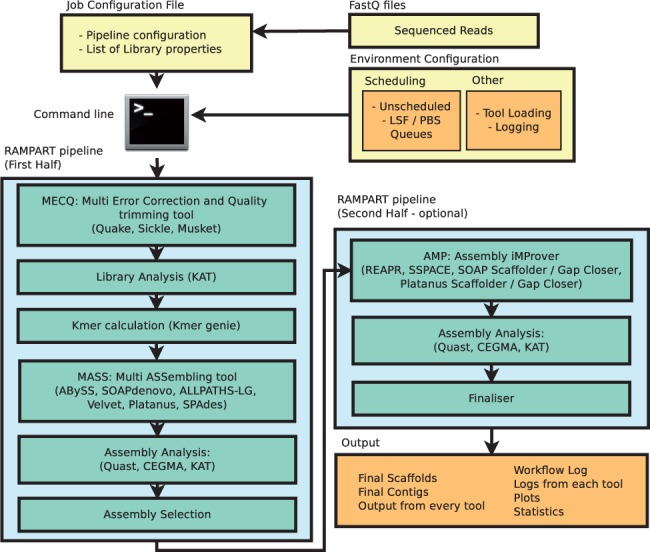
![PDF] RAMPART: a workflow management system for de novo genome assembly | Semantic Scholar PDF] RAMPART: a workflow management system for de novo genome assembly | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/b613fe5a2e74f26cea1065c75a0f93d20fb11c54/2-Figure1-1.png)